Write/modify a simple python script
- Status: Closed
- Prize: $30
- Entries Received: 2
- Winner: gpmember88
Contest Brief
Dear all,
I need to edit my python script (or create a new one) to analyze multiple sequence of proteins:
I have two sets of data (see attached examples):
1. XXX_clustallo.fasta files (contain aligned protein sequences)
2. XXX.csv files (contain information about organisms from which protein sequences in XXX_clustallo.fasta files are derived)
I need to figure out which of the protein sequences are mutated, for which I wrote a python script (script.py in the attachment).
If I type the following:
python script.py LeuRS T1524,D1527,RK1528
#(LeuRS and T1524,D1527,RK1528 are variable)
then:
1. The script finds a file called LeuRS_clustallo.fasta
2. Then, the script reads each line (each line corresponds to one protein sequence) in LeuRS_clustallo.fasta and verifies:
whether position 1524 in this line is represented by letter "T",
whether position 1527 in this line is represented by letter "D",
whether position 1528 in this line is represented by letter "R" or letter "K"
3. Finally, this script uses XXX.csv file to create XXX_new.csv according to the following rule:
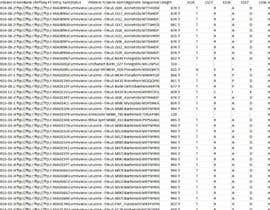
A. The script creates a new column called "mutation status" in the XXX_new.csv file.
B. If a line (a protein sequences) in XXX_clustallo.fasta file carries letters T at 1524, D at 1527 and R or K, then the script will type "non-mutated" in the cell column of the .csv file
C. If a line (a protein sequences) in XXX_clustallo.fasta file carries at least on letter that is not T at 1524, or not D at 1527, or not R or K at 1528, then the script will type "mutated" in the corresponding cell of the .csv file
(As you can see from the code, to find the correspondence between lines of .csv file and lines of .fasta file, my script compares values from the "Entry name" column in .csv file and identical values in the header of each line in the .fasta file. )
I need to modify my script to add an additional information to the XXX_new.csv file:
D. Create additional columns (in my example, these columns should be called 1524, 1527 and 1528)
E. Fill these columns with the actual symbol at this position in each protein
F. Create column "Total number of mutations" that will show the total of mutations in each protein sequence. For instance, if a protein sequence has letter T at position 1524, D at position 1527 and R or K at position 1528, then the corresponding number will be "0" (no mutations). And if the protein has letter Y at 1524, E at 1527 and - at 1528, then the corresponding number will be "3" (all three residues are mutated or missing).
I hope my description is clear. Your questions are very welcomed.
This is a one-time task, but I will need help with similar tasks in the coming two weeks. So, I'm looking to find someone who can also help me in the future.
If you manage to write a script, could you make a screenshot of LeuR_new.csv file to show additional columns and their values for a couple of the protein sequences?
Thank you for your time and effors,
Sergey
Recommended Skills
Employer Feedback
“It was my pleasure working with GP and I will definitely consider cooperating with him in the future. Actually, I was amazed by how well and quickly he understands the task and how little time he needs to complete it. Amazing pro!”
![]() sergeyvmelnikov, United States.
sergeyvmelnikov, United States.
Top entries from this contest
-
gpmember88 Israel
-
Vijayabhaskar96 India
Public Clarification Board
How to get started with contests
-
Post Your Contest Quick and easy
-

Get Tons of Entries From around the world
-

Award the best entry Download the files - Easy!